INTRODUCTION
Stanniocalcin (STC) is a glycoprotein hormone that was initially identified in fish (Wagner et al., 1986). Subsequent studies have identified STC in the mammalian genome, including a homologous protein, STC2 (Chang and Reddel, 1998; Ishibashi et al., 1998; Morita et al., 1999). In mammals, STC2 is expressed in multiple tissues, including the kidney, brain, and reproductive organs and functions in a paracrine or autocrine fashion (Chang et al., 1995; Ishibashi and Imai, 2002; Luo et al., 2005). STC2 has been implicated in various physiological processes, including cellular growth, metabolism, and the stress responses (Ito et al., 2004; Low and Wong, 2010; Chen et al., 2016).
STC2 is also abundantly expressed in skeletal and cardiac muscle (Ishibashi et al., 1998). STC2 is involved in the regulation of insulin-like growth factor (IGF) 1 through the control of pregnancy-associated plasma protein-A (PAPPA) (Jepsen et al., 2015). In a previous study, STC2 knock-out mice exhibited increased muscle mass which was accompanied by elevated level of IGF axis components and IGFBP4 in overloaded muscles of STC2 knock-out mice (Lionikas et al., 2023). STC2 was reported to inhibit growth and neonatal viability in transgenic mice overexpressing human STC2, suggesting that STC2 can reduce endochondral bone development and skeletal muscle growth (Gagliardi et al., 2005). A genome-wide association study (GWAS) on hindlimb skeletal muscle mass identified STC2 as a modifier of myogenesis in humans and mice, and knocking-down of STC2 increased the length of myotubes (Cordero et al., 2020). On the other hand, Mittapalli et al. (2006) reported that STC2 expression is delayed compared to STC1, which is expressed in myoblasts prior to differentiation, suggesting that STC2 may play a role in differentiating muscle cells and fibers. Despite extensive studies on the involvement of STC2 in muscle biology, the precise mechanisms and specific roles of this protein remain unclear.
Myogenesis, the process of muscle formation through the fusion of myoblasts, involves the regulation of various genes and is a complex and essential process for muscle growth, repair, and maintenance (Kim et al., 2015). Understanding the functions and mechanisms of genes involved in muscle formation can have significant implications for preventing muscle-related diseases, reducing production costs, and advancing genetic research and biotechnology to improve the livestock production efficiency. This study aims to explore the functional role of STC2 in poultry muscle formation by overexpressing STC2. Unlike previous studies, we investigated how STC2 regulates the expression of various genes involved in myogenesis. This study will contribute to new insights into the molecular function of STC2 in muscle cells.
MATERIALS AND METHODS
The coding sequence of quail STC2 (GenBank accession no.: XM_015876362.2) was PCR-amplified and inserted into the multicloning site of the pcDNA3.1 expression vector (Invitrogen, Grand Island, NY, USA). A hemagglutinin (HA) tag sequence was appended just before the stop codon of STC2 gene. Finally, the STC2 sequence was confirmed by sequencing.
QM7 cells (obtained from the American Type Culture Collection, Manassas, VA, USA) were cultured in a growth medium consisting of Medium 199 supplemented with 10% fetal bovine serum (FBS; Gibco, Grand Island, NY, USA), 1% chicken serum (CS; Sigma-Aldrich, St. Louis, MO, USA), and 1% antibiotic-antimycotic (ABAM; Gibco, Waltham, MA, USA). These cells were maintained in an incubator at 37°C in a CO2 atmosphere. To prevent differentiation, the cells were subcultured before reaching confluence. For differentiation, the growth medium was replaced with a differentiation medium composed of Medium 199 with 0.5% FBS, 0.1% CS, and 1% ABAM. Fourteen 35-mm dishes were seeded with 6 × 105 cells one day prior to transfection. Transfection with STC2-containing or empty expression vectors (EV) was performed using jetOPTIMUS® DNA transfection reagent (Polyplus, Illkirch, France) following the manufacturer's instructions. Differentiation began the following day, and the differentiation medium was replaced with fresh medium on day 2.
Differentiation was assessed at four specific time points: day 0, marking the initiation of differentiation; day 1, representing the transition in myogenic gene expression; day 2, indicating the active development of myotubes; and day 4, signifying the completion of myotube formation. mRNA samples were collected at all four time points (days 0, 1, 2, and 4) or solely on day 4. Protein samples were taken at two time points (days 0 and 4), while immunofluorescence samples were collected exclusively on day 4. At each designated time point, samples were carefully prepared for subsequent analysis.
For immunofluorescence staining, the cells were washed with phosphate-buffered saline (PBS) and then fixed with 10% neutral formalin for 15 minutes. Subsequently, the cells were permeabilized using 0.3% NP-40 for 20 minutes and blocked with 5% non-fat dry milk in PBS containing 0.1% Tween-20 (PBST) for 30 minutes. The cells were then stained with the primary antibody, anti-myosin heavy chain (MyHC) (MF20; Developmental Studies Hybridoma Bank, Iowa City, IA, USA), for 1 hour. Following this, anti-mouse immunoglobulin G (IgG) conjugated with CruzFluorTM594 (Santa Cruz Biotechnology, Dallas, TX, USA) was used as the secondary antibody for 1 hour. Non-fat dry milk was reduced to 1% during antibody treatment. The cell nuclei were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) for 5 minutes in PBS. Images of the stained cells were captured using an inverted fluorescence microscope (CKX53; Olympus, Tokyo, Japan). The lengths of the myotubes were measured in MyHC-positive cells.
Total proteins were extracted using 1× lysis buffer. The extracted proteins were then mixed with an equal volume of 2× Laemmli sample buffer (Bio-Rad, Hercules, CA, USA) containing β-mercaptoethanol and boiled at 100°C for 5 minutes. Protein quantification was performed using Coomassie staining. The protein samples were separated by polyacrylamide gel electrophoresis and transferred onto a polyvinylidene fluoride (PVDF) membrane. The membrane was blocked with 5% non-fat dry milk in TBST for 1 hour and then incubated overnight at 4°C with anti-HA tag (Santa Cruz Biotechnology, Dallas, TX, USA) or anti-MyHC (NA4; Developmental Studies Hybridoma Bank, Iowa City, IA, USA) antibodies. The next day, the membrane was treated with goat anti-mouse IgG-HRP secondary antibodies (Thermo Fisher, Carlsbad, CA, USA) for 1 hour at room temperature. During antibody treatments, non-fat dry milk was reduced to 2.5%. After applying ECL solution, images were captured using ImageQuantTM LAS 500 (Cytiva).
Total mRNA was extracted from the samples obtained on days 0, 1, 2, and 4 of differentiation using RNAiso plus (Takara Bio Inc., Shiga, Japan). The quality of the isolated RNA was confirmed using a P200 Micro-volume spectrophotometer (Biosis Design, Gwangmyeong, Republic of Korea) and electrophoresis. Using 1 μg RNA, cDNA synthesis was performed with the DiaStar RT kit (SolGent, Daejeon, Republic of Korea) according to the manufacturer’s instructions. qRT-PCR was conducted using the Bio-Rad CFX ConnectTM Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). Primer pairs designed for qRT-PCR are shown in Table 1. The single amplicon of each primer set was confirmed by a melting curve analysis. Target genes were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH). The expression levels of the target genes were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001).
All experiments were independently conducted at least three times. Data are presented as the mean ± standard error of the mean (SEM) and were analyzed using Student’s t-test and two-way analysis of variance (ANOVA) in the R package (R Foundation for Statistical Computing, Vienna, Austria). For multiple groups showing significance in the two-way ANOVA test, post-hoc comparisons were conducted using the least significant difference (LSD) test. Statistical significance was defined as P<0.05.
RESULTS
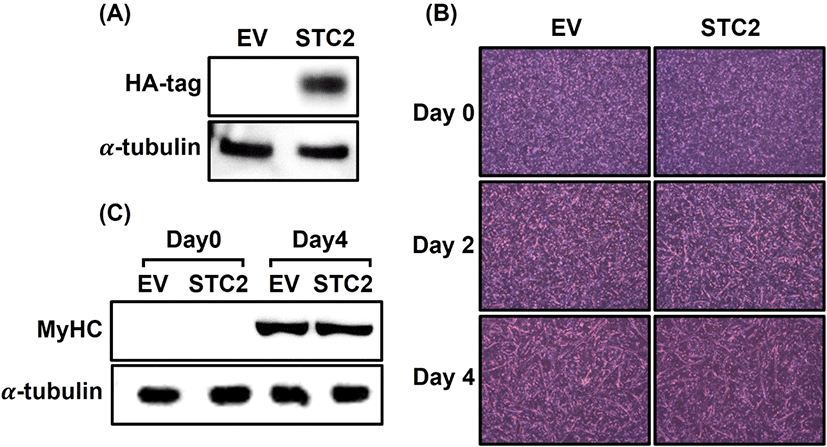
To explore the function of STC2 in muscle formation, an STC2 overexpression vector was transfected into QM7 cells. Since no antibody was specifically designed to detect quail STC2 protein, the overexpression of STC2 was confirmed by detecting the HA-tag with an HA antibody on day 0 of differentiation in STC2-overexpressing (OE) cells (Fig. 1A). In contrast, HA-tag expression was not detected in the control cells. Myotube formation in STC2-OE cells was then assessed in comparison to control cells (Fig. 1B). Initially, no differences were observed between the groups at the start of differentiation. However, STC2-OE cells showed a quicker initiation of myogenesis compared to the control cells on day 2. On day 4, when sufficient differentiation was achieved, noticeable differences were evident: the elongation of myotubes was greater in STC2-OE cells than in the control cells. Additionally, STC2-OE cells exhibited a higher occurrence of interconnected myotube formations compared to the control cells. To investigate the degree of differentiation due to STC2 overexpression, the amount of MyHC protein was compared between the groups on day 0 and 4 (Fig. 1C). In both groups, MyHC protein was not detected on day 0. On day 4, MyHC expression levels were similar between the groups.
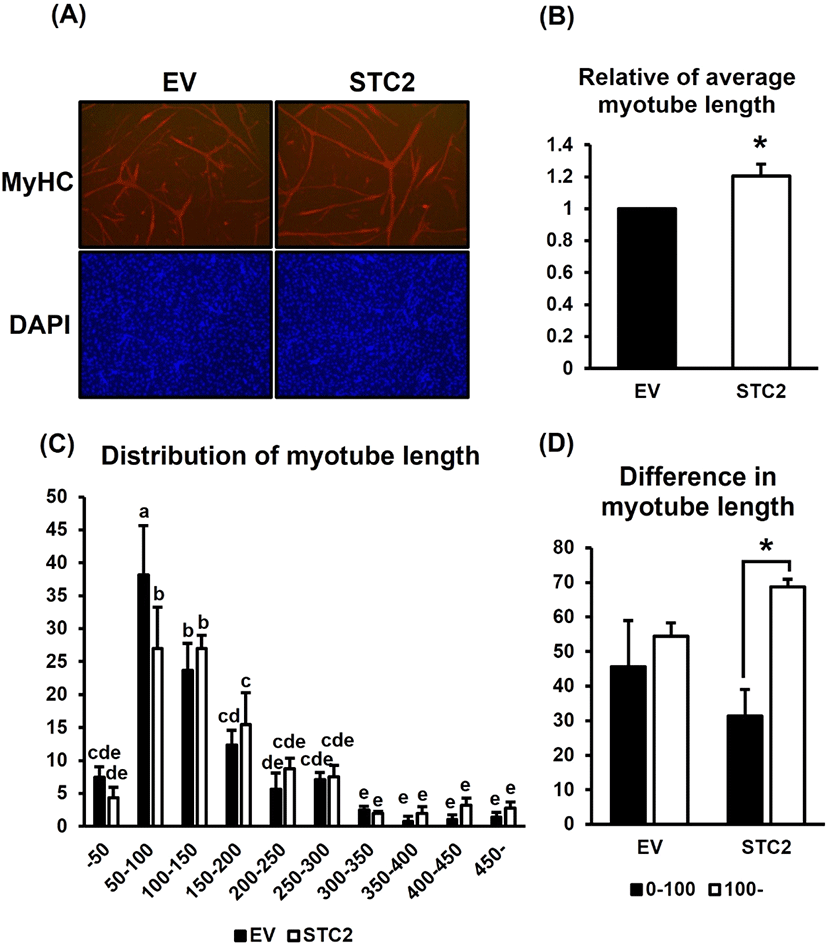
To further verify the differences in the formed myotubes, the cells were stained with anti-MyHC and DAPI, and the myotube length was subsequently measured in each group (Fig. 2). STC2-OE cells formed longer myotubes compared to the control cells (Fig. 2A). Quantitatively, the myotube length in STC2-OE cells increased by 20% compared to the control (P<0.05) (Fig. 2B). When measuring the frequency of myotube lengths, a tendency toward longer myotubes was observed in STC2-OE cells with a significant difference compared to the control in the range of 50—100 (Fig. 2C). Myotube length was also measured based on specific length ranges to investigate differences in myotube length (Fig. 2D). In contrast to control cells, STC2-OE cells formed a significantly greater number of longer myotubes (P<0.05). These results suggested that STC2 does not affect the overall amount of muscle formation; however, it may play a role in regulating the elongation of myotubes during myogenesis.
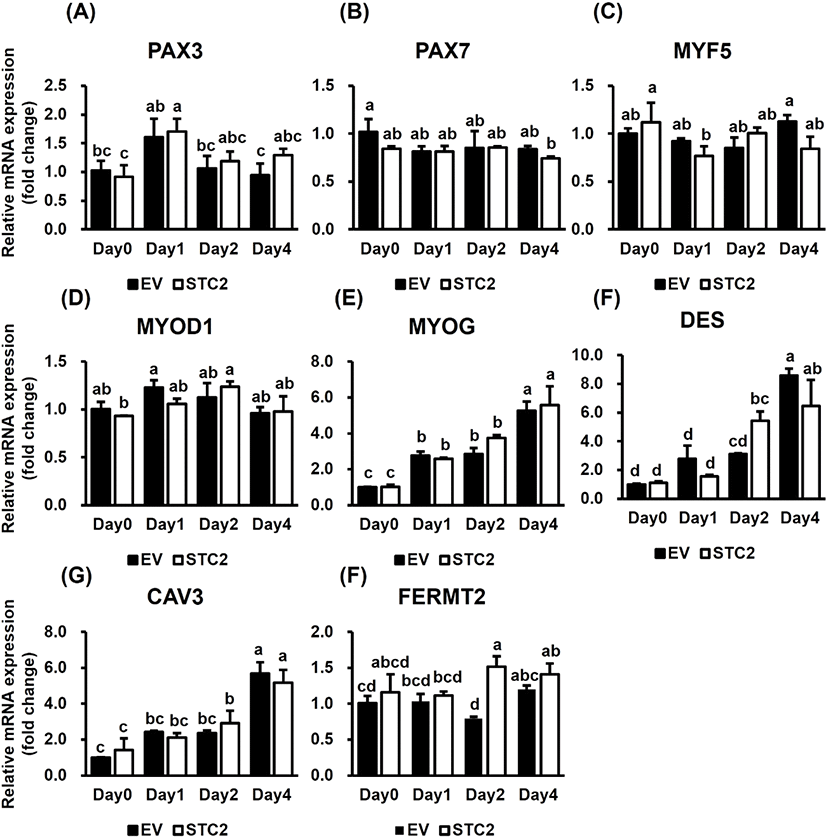
To assess the role of STC2 on the expression of myogenic regulatory factors and genes related to muscle differentiation and maturation, qRT-PCR was conducted (Fig. 3). Paired box 3 (PAX3) exhibited significant changes in expression during differentiation in both groups (P<0.05), although there were no differences between the groups (Fig. 3A). Similarly, paired box 7 (PAX7), myogenic factor 5 (MYF5), and myogenic differentiation 1 (MYOD1) showed no significant differences between the groups (Fig. 3B, 3C, and 3D). Myogenin (MYOG), Desmin (DES), and caveolin 3 (CAV3) gradually increased during myogenesis in both groups (P<0.001); however, there were no significant differences between the groups (Fig. 3E, 3F, and 3G). FERM domain containing kindlin 2 (FERMT2) was upregulated in STC2-OE cells on day 2 compared to the control cells (P<0.01) (Fig. 3H). These data indicated that STC2 regulates FERMT2 expression during myogenesis.
DISCUSSION
In this study, the role of STC2 in muscle formation and myotube elongation was investigated by overexpressing STC2 in QM7 cells. As a result, STC2-OE cells initiated myotube formation more rapidly on day 2 compared to the control group, and on day 4, STC2-OE cells showed significantly longer myotubes. Quantitative analysis revealed that STC2-OE cells produced approximately a 20% increase in myotube length and formed longer myotubes than short ones compared to control cells. Despite these structural changes, the overall amount of MyHC protein, a key marker of muscle differentiation (Emerson and Bernstein, 1987), was similar between the groups on day 4. A previous study has shown that STC2 enhances osteoblast differentiation (Zhou et al., 2016) and that myoblast cell lines have the capability to differentiate into osteoblasts (Yaffe and Saxel, 1977; Huang et al., 2012). Additionally, STC2 expression observed after myoblast differentiation in skeletal muscle, suggests that STC2 plays a crucial role in the differentiation of muscle cells and myofibers (Mittapalli et al., 2006). This study suggests that while STC2 plays an important role in promoting myocyte development during the later stages of myogenesis, it primarily influences myotube length without significantly affecting the overall quantity of muscle formation.
This study also confirmed a molecular mechanism involving the correlation of STC2 with muscle-related genes. The expression of paired box genes and myogenic regulatory factors (MRFs), which regulate myogenic development and differentiation (Zanou and Gailly, 2013; Buckingham and Relaix, 2015), did not differ between the groups. Similarly, DES and CAV3, markers of muscle cell maturation (Vanderven et al., 1992; Schöneich et al., 2014) were no significant differences in the expression of these genes between the groups. However, there were elevated during differentiation in both groups, maintaining muscle structural integrity. A previous study reported that STC2 does not affect cell proliferation or cell size during pre- and postnatal growth restriction, suggesting that it may influence developmental programming (Gagliardi et al., 2005). Therefore, this study suggested that while STC2 does not regulate muscle-related genes, it may influence cell morphological characteristics during muscle formation.
Although STC2 did not have a broad effect on MRF expression, it may play a role in regulating certain aspects of muscle differentiation, such as FERMT2 expression. FERMT2, also known as Kindlin2, is involved in cell adhesion and signaling pathways (Megeney and Rudnicki, 1995). Since myoblast fusion was disrupted by the FERMT2 knockdown, it has been demonstrated that FERMT2 is essential for myoblast fusion (Suzuki et al., 2015). Additionally, Dowling et al. (2008) demonstrated that FERMT2 is essential for myocyte elongation. Interestingly, in this study, FERMT2 was significantly upregulated in STC2-OE cells on day 2. The upregulation of FERMT2 aligns with the observed increase in myotube length, indicating that STC2 may influence specific pathways associated with cell adhesion and signal transduction, which are crucial for muscle cell fusion and elongation.
In conclusion, STC2 may increase myotube elongation primarily through the regulation of genes such as FERMT2. Specifically, STC2 is suggested to regulate cell fusion and length during the mid- and late stages of myocyte development. However, further studies are necessary to elucidate the detailed signaling pathways and molecular interactions mediated by STC2 in muscle differentiation, particularly those related to cell fusion and other key mechanisms.
SUMMARY
Stanniocalcin 2(STC2) 유전자는 다양한 세포 유형에서 세포 성장, 대사, 스트레스 반응을 포함한 여러 생리적 과정을 조절한다. STC2는 골격근에서 풍부하게 발현되지만, 골격근 분화에서의 구체적인 역할과 기능은 아직 완전히 이해되지 않았다. 본 연구에서 근형성 동안 STC2의 역할과 조절 기전을 조사하기 위해 STC2 발현 벡터를 메추리 근육 세포 주에 형질 도입하였다. STC2 과발현 세포는 대조군 세포와 비교하였을 때, 형태적으로 더 긴 근관을 형성했으며 상대적으로 근관의 길이가 증가하였다. 또한, STC2 과발현 세포에서 더 많은 핵을 가진, 긴 근관을 형성하였다. STC2가 근육 분화에 미치는 기전을 조사하기 위해 근육 분화와 성숙을 조절하는 것으로 알려진 여러 유전자의 mRNA 발현 수준을 비교하였다. 이러한 유전자들 중에, 대조군 세포와 비교하여 STC2 과발현 세포는 Fermitin family member 2 (FERMT2)의 발현이 분화 2일 차에 유의하게 증가하였다. STC2는 근관의 융합에 필수적인 FERMT2 발현을 매개하여 근관의 길이를 조절할 것이라고 사료된다. 본 연구는 STC2가 근형성에의 역할에 대한 중요한 정보를 제공하며, 이는 근육 관련 질병의 치료 및 가금류 생산 향상에 도움이 될 수 있을 것으로 사료된다.








