INTRODUCTION
Infectious bursal disease (IBD), an acute, highly contagious, and immunosuppressive disease in young chickens, causes considerable economic losses to the poultry industry. Infectious bursal disease virus (IBDV) destroys B-lymphocytes in the bursa of Fabricius of young chickens, inducing immunosuppression (Jackwood, 2017; Saif, 1991). IBDVs are classified under the Avibirnavirus genus of the Birnaviridae family, have a non-enveloped icosahedral capsid with a double-stranded RNA (dsRNA) genome with two segments (segments A and B). Segment A is approximately 3.2 kb and contains two open reading frames (ORFs) that encode viral proteins including VP5, VP2, VP4 and VP3 (Fahey et al., 1989; Bayliss et al., 1990). VP2 is the major structural protein, which is involved in antigenicity, cell tropism, virulence and apoptosis (Coulibaly et al., 2005). The amino acid (aa) residues 206−350 of VP2 have been identified as the hypervariable region (HVR) which contains four hydrophilic regions, including aa 210−225 (peak A), 247−254 (minor peak 1), 281−292 (minor peak 2), and 312−324 (peak B) (Letzel et al., 2007). Segment B is approximately 2.8 kb and encodes VP1, an RNA-dependent RNA polymerase (RdRp), which plays an important role in viral replication and genetic evolution (Escaffre et al., 2013).
IBDVs are divided serotypes I and II. Serotype I IBDVs are pathogenic to chickens and are further classified into seven subtypes, classical / attenuated IBDV (cv/at IBDV, G1), antigenic variant IBDV (avIBDV, G2), very virulent IBDV (vvIBDV, G3), distinct (d) IBDV (dIBDV, G4), variant / classical recombinant (G5), dIBDV from Italy (G6) and Australia (G7) (Jackwood et al., 2018). Serotype II IBDVs have been isolated from turkeys and are nonpathogenic to chickens (Ismail et al., 1988).
IBDV was first isolated in 1957 in the United States (Cosgrove, 1962). Subsequently vvIBDV and avIBDV have emerged (Chettle et al., 1989; Jackwood and Saif, 1987). Over the past 30 years, vvIBDV with an extremely high mortality rate has caused considerable economic losses to the poultry industry (Jackwood, 2017). Intermediate or intermediate plus live attenuated vaccines and inactivated vaccines were widely used to control IBDV infection, and vvIBDV is well contained worldwide. However, in recent years, novel avIBDV that emerged in China rapidly spread to other countries (Fan et al., 2019; Thai et al., 2021; Xu et al., 2020). avIBDV has no mortality but causes severe atrophy of the bursa of Fabricius and induces severe immunosuppression in chickens (Fan et al., 2019; Xu et al., 2020). In South Korea, the first IBD outbreak was reported in 1980 and four genogroups of IBDV (G1-G4) have been isolated to date (Jeon et al., 2009; Kim et al., 2010; Kwon et al., 2000). In the present study, we focused on the genetic and pathogenic characterization of the avIBDV from chickens in Korea. This study provides information about genetic and pathogenic characterization of currently circulating avIBDV in Korea.
MATERIALS & METHODS
Bursal samples were collected between 2019 and 2020 from suspected IBDV-infected chickens from chicken farms (Jeonbuk and Gyeongki provinces). Bursal samples were homogenized in 10% (w/v) phosphate buffered saline [PBS, pH 7.4; supplemented with 100× antibiotic−antimycotic (Gibco, New York, USA)]. The homogenates were centrifuged at 3,000×g for 10 min at 4°C, and the supernatant was collected for subsequent IBDV detection and analysis. The supernatant was then conserved in aliquots at −70°C for virus isolation and viral RNA extraction.
Viral RNA was extracted from the clarified bursal samples using the 5X MagMAX™ − Pathogen RNA/DNA kit (Thermo Fisher Scientific, Vilnius, Lithuania) with KingFisher Duo Prime Purification system (Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s protocol. Viral cDNA was generated from RNA samples using GoScript™ reverse transcriptase (Promega, Madison, WI, USA) with random primers (9‐mers; TaKaRa Bio. Inc., Otsu, Shiga, Japan). In the reverse transcription (RT) reaction, 8 μL of extracted RNA and 2 μL of dimethyl sulfoxide (Tedia, Ohio, Fairfield, USA) were heated at 100°C for 5 min and then placed in an ice bath for 5 min. Then, the following materials were added to this reaction mixture: 8 μL of GoScript™ 5X RT reaction buffer (Promega, Madison, WI, USA), 10 μL of 2.0 mM of each dNTP (SolGent, Daejeon, Korea), 4 μL of MgCl2 (Promega, Charbonniere, France), 1 μL of 20 units Recombinant RNasin® Ribonuclease Inhibitor (Promega, Madison, WI, USA), 1 μL of GoScript™ reverse transcriptase, 1 μL of 50 pmol random primer, and 4 μL of diethylpyrocarbonate-treated water (DEPC water; Biosesang, Seoul, Korea); a final volume of 39 μL was obtained. The RT reaction mixture was incubated in this sequence: 25°C for 5 min, 42°C for 60 min, and 70°C for 15 min to inactivate the enzyme (Kim et al., 2022). The RT-PCR was performed using the forward primer IBDV-F1 (5′-GCCCAGAGTCTACACCAT-3′) and the reverse primer IBDV-R1 (5′-CCCGGATTATGTCTTTGA-3′), which amplified 743-bp fragment covering the hypervariable region (HVR) of the VP2 gene (Jackwood et al., 2006). The RT-PCR products were purified and the nucleotide sequences assembled using an ABI 3730XL DNA Analyzer (Applied Biosystems, Foster City, CA, USA) by SolGent (Daejeon, Korea).
Based on the nucleotide and deduced amino acid sequences of the VP2 HVR, phylogenetic analysis was conducted. Sequence alignments were performed on the nucleotide sequences using GENETYX software (Ver 6.1; Genetyx Corp., Tokyo, Japan). A 360-bp fragment (661−1,021 nt) of VP2 HVR was aligned, and phylogenetic trees were constructed using the neighbor-joining method and 1,000 bootstrap replications with the MEGA X software. (MEGA, 2018). The GenBank accession numbers and the nation of the reference strains used in this study are provided in Table 1.
Bursal homogenates were inoculated into 10-day-old specific pathogen-free (SPF) embryonated chicken eggs via the chorioallantoic membrane (CAM) route. Embryonated eggs were observed daily for 5 days by candling. At 5 days post-inoculation (dpi), CAM of the infected embryos were harvested and homogenized by freezing and thawing three times; then, 1.5 mL PBS was added, followed by centrifugation at 3,000×g for 10 min. The 50% embryo infectious dose (EID50) was calculated by the method of Reed and Meunch (Reed & Muench, 1938).
RESULTS
Four avIBDV strains were isolated and confirmed by RT-PCR. Details of these strains isolated from chicken farms are summarized in Table 2. Chickens originated from three broiler farms and one Korean native chicken farm. Most farms were reported to show low mortality rate but were co-infected with viral diseases such as inclusion body hepatitis (IBH) and infectious bronchitis (IB) and bacterial diseases such as Colibacillosis and Clostridial disease. Among these strains, two strains were isolated from IBDV-vaccinated chicken farms.
Phylogenetic analysis of VP2 HVR nucleotide sequences of reference strains showed that these strains were divided into seven major branches. The antigenic variant group (G2) was divided into four-sublineages (G2a, G2b, G2c and G2d). According to the phylogenetic analysis, all four strains in the present study were classified into one branch with the reference strains of the antigenic variant group (G2). A19-MRA-016 was clustered with the representative avIBDV strains from G2a lineage which were demonstrated in America and Korea (V1, and 09D243 strains, respectively). A20-ETC-014, A20-CFR-001 and A20-MRA-149 were clustered G2d lineage together with SHG19 and ZD-2018-1 which recently emerged in China. A05-HL-144, which was used in pathogenicity experiment was clustered in the dIBDV group (G4) (Fig. 1).
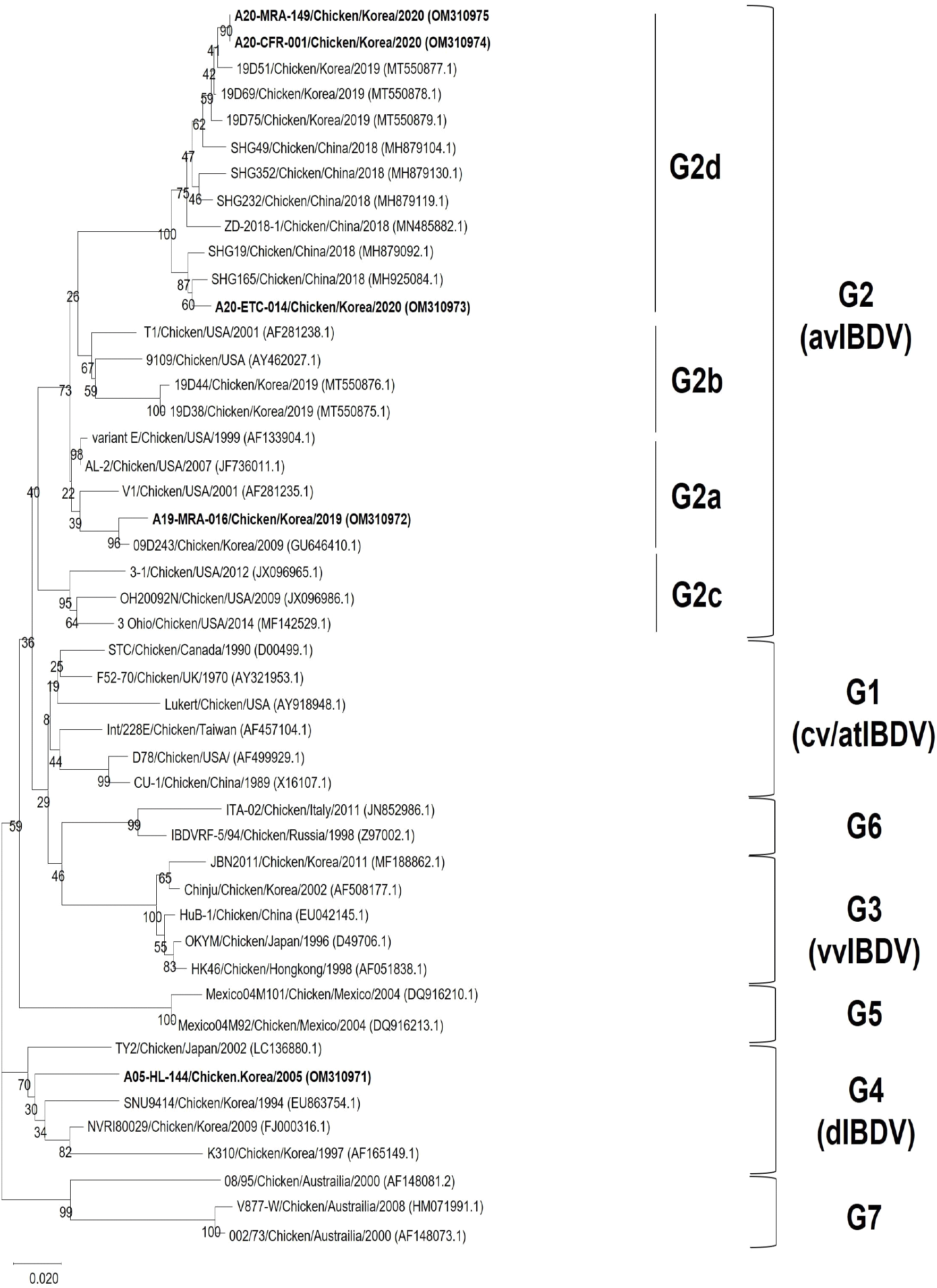
The variant strains in the present study were classified into two lineages: G2a and G2d; and these strains showed obvious differences in VP2 gene sequence. A19-MRA-016 was more closely related to variant strains from G2a and G2b lineages (93.6−98.3% nucleotide sequence identity) than the G2d lineage (91.9−92.5% nucleotide sequence identity). Additionally, A20-ETC-014, A20-CFR-001, and A20-MRA-149 showed 96.1−100% nucleotide sequence identity within the G2d, which was higher than the identity to the previous lineages in G2a (91.9−93.6%, nucleotide sequence identity) (Table 3).
Further molecular characterization of avIBDVs was conducted by multiple alignments of the deduced amino acid sequences of HVRs of the VP2 gene (221 to 340 aa). All the variant IBDV strains possessed similar typical amino acid residues 222T, 249K, and 286I. Notably, two conserved amino acid residues including 252I, and 299S were only existed in the Chinese variant IBDVs (G2d), which are distinctly different from the American variants (G2a) (Table 4).
DISCUSSION
Infectious bursal disease (IBD) is an acute, highly contagious and immunosuppressive disease in young chickens, causes considerable economic losses to the poultry industry (Van den Berg et al., 2000). Since the first emergence of classic IBDV in the USA was documented in 1957, and avIBDV was reported in the USA in 1987, which evaded the immune protection of classic IBDV (Cosgrove, 1962; Jackwood & Saif, 1987). However, over the past 30 years, avIBDV was neglected because of the subsequent cases of vvIBDV, which has high mortality and morbidity with economic losses (Jackwood et al., 2006). However, the novel avIBDV emerged in 2019 in China (Fan et al., 2019). This avIBDV has a low mortality, but it causes severe atrophy of bursa, which induces immunosuppression and avIBDV-infected birds are susceptible to secondary infection of bacterial and viral diseases such as IB, IBH, colibacillosis (Berg, 2000; Jackwood, 2017; Xu et al., 2020).
In our study, four avIBDVs were detected in Korean broilers and native chickens. Among the four avIBDVs, two were isolated from chicken farms immunized with the intermediate-plus IBDV vaccine. However, gross lesions such as atrophy and inflammation in these birds showed no differences as compared to non-vaccinated chickens. Also, three isolates were co-infected with other viral and bacterial diseases. In previous studies, the commercial vaccines against avIBDV may not have provided complete protection to chickens (Fan et al., 2019). Therefore, it is necessary to evaluate the efficacy evaluation of the avIBDV of the current commercial vaccine, and continuous surveillance of the avIBDV is also needed. In addition, it is imperative to study the immunosuppressive properties of the avIBDV and the risk of co-infection with other viral and bacterial diseases.
VP2 gene is a capsid protein of IBDV and includes a P domain. The P domain is involved in antigenicity and host cell adhesion. The HVR of the VP2 gene is frequently used for genetic analysis of IBDV (Coulibaly et al., 2010; Jackwood et al., 2018). All four isolates in this study belonged to the antigenic variant group (G2). Three of the isolates belonged to the G2d group, which is a novel avIBDV that recently occurred in China, and one of isolates belonged to the G2a group, similar to the American variant strain. A novel antigenic variant IBDV was first reported in China in 2019, and is also being detected in Korea, Japan, and Malaysia (Aliyu et al., 2021; Fan et al., 2019; Myint et al., 2021; Thai et al., 2021). In Korea, the avIBDV 09D243 similar to the Variant E was first reported in 2010 (Kim et al., 2010). A total of 5 isolates were reported in 2021, including two isolates were in the G2b group and three were in the G2d group (Thai et al., 2021). In addition, although viruses such as K310, NVRI80029, and SNU 9414, similar to A05-HL-144 used in this study, were reported as Korean antigenic variants, these strains were regarded as dIBDV and classified as genotype 4 (Jeon et al., 2009; Kwon et al., 2000; Tomás et al., 2019). Furthermore, the Chinese variant IBDVs (G2d) were obviously different from the American variant IBDVs (G2a, G2b). A20-ETC-014, A20-CFR-001, and A20-MRA-149 showed 96.1−100% sequence identity with the G2d, which was higher than the identity to the previous other lineages (91.9−93.6%). Therefore, it is important to study the transmission of avIBDV, and to study the effect of the genome mutations in antigenic variant strains on the pathogenicity in the chicken.
As a result of amino acid (aa) analysis of VP2 gene HVR, characteristic amino acids, including 222T, 249K, and 286I mutations were identified. In the case of 222 (aa), it changes the reactivity of monoclonal antibodies and induces immune evasion (Jackwood & Sommer-Wagner, 2011; Michel & Jackwood, 2017). In the case of 249 (aa), it was confirmed that the virulence was weakened when the Q249R mutation occurred, providing evidence related to virulence (Qi et al., 2013).
Compared with the American antigenic variant IBDV, distinct amino acid residues in VP2 (252I and 299S) were observed in the Chinese variant IBDVs (Fan et al., 2019). In the case of 299S, it is both a characteristic amino acid appearing in antigenic variants and one of the characteristic amino acids of very virulent IBDV. The biological role of these amino acids is unknown. Therefore, it is necessary to study the effect of amino acid mutations on pathogenicity (Jackwood et al., 2018).
In conclusion, we reported the co-occurrence of different sub-lineages of avIBDVs in South Korea. In addition, we show the possibility that avIBDVs cannot be controlled using the current anti-IBDV vaccine, considering their continuous occurrence in vaccinated flocks. Therefore, it is necessary to evaluate the efficacy of current commercial vaccines and pathogenicity tests.
SUMMARY
전염성 F낭병은 닭에서 F낭의 위축 및 염증이 나타나고, 이로 인해 면역억제를 특징으로 하며, 급성, 높은 점염성을 가지는 질병이다. VP2 유전자의 염기서열 분석을 통해 유전형이 분류되고, 유전형에 따라 항원성 및 병원성에 큰 차이를 나타내고 있다. 최근 중국에서 발생한 항원 변이주의 경우, 기존의 미국형 항원 변이주와는 다른 분자 특성을 보이는 것으로 나타났다. 최근 국내에서도 이러한 중국형 항원 변이주가 발생하고 있고, 따라서 본 연구에서는 국내 양계농장에서 분리된 항원 변이주의 유전체 분석을 통한 분자적 특성을 확인하였다. 국내 육계 및 토종닭으로부터 4개의 항원 변이주를 RT-PCR을 통해 확인하였고, chorioallantoic membrane 접종을 통해 분리하였다. VP2 유전자의 hypervariable region의 분석 결과, 4개의 항원 변이주 중 1개는 미국형 항원 변이주로 확인되었고, 이는 2009년 국내에서 보고된 09D243 균주와 유사한 것으로 나타났으며, 3개의 중국형 항원 변이주는 최근 국내 및 중국의 유행주와 유사한 것으로 확인되었다. 우리의 결과에서 국내 양계농장에서 중국형 및 미국형 항원 변이주가 퍼져 있음을 확인하였고, 또한 백신을 접종한 농장에서의 발생도 이뤄지고 있어 항원 변이주에 대한 상용화 백신의 보호 효능에 대한 연구도 필요합니다.








